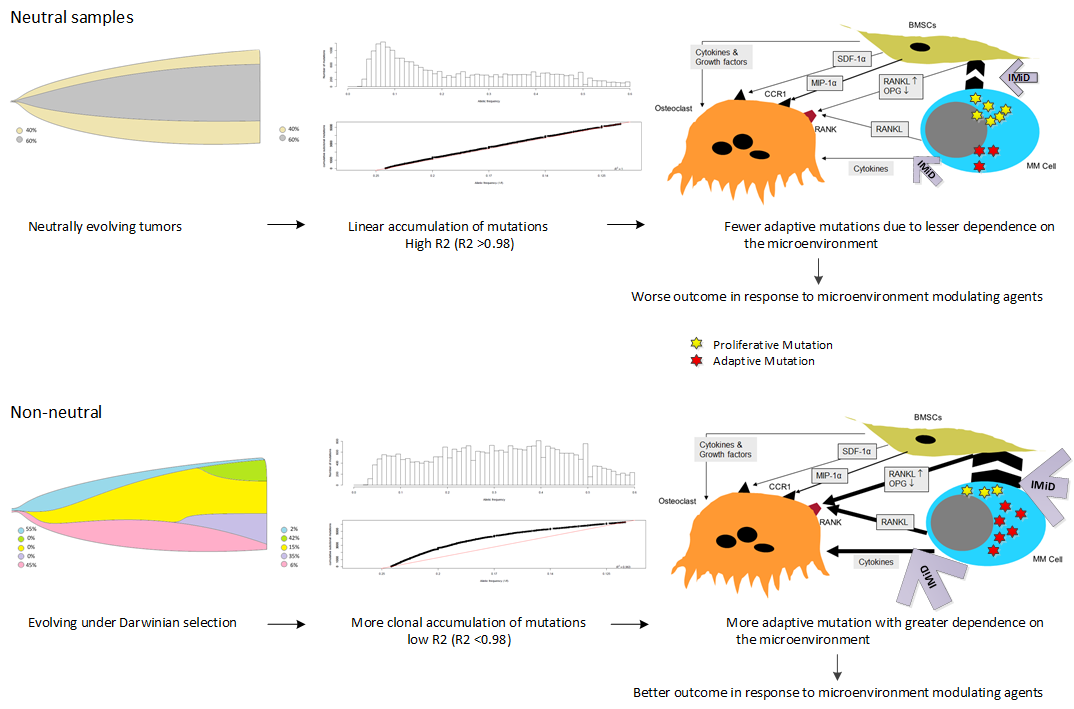
Neutral Evolution
Background
An ICR colleague resurrected the evolutionary theory of neutrality and measured the concept through bulk
sequencing data. Sottoria had shown that some tumours are driven by such strong initial drivers, that they evolve neutrally and are more resistant to forming sub-clones
in response to selectional pressure from the environment. Sottoriva suggested that tumour neutrality could have prognostic importance, we were able to be the first to
investigate this. The approach by
Sottoriva et al has been critiqued
in the literature. The practical problem is the influence of somatic copy number changes and the stringency of the R2 cut-off. It is likely that R2
needs to be assessed for each tumour type, rather than a definitive threshold for all tumours. The usefulness of this method for clinical predication is questionable, as
tumour neutrality is measured during a particular time span and changing the environment could change a tumour neutral tendency. So that neutral evolution status is a moving target,
and clinicians would be better served taking serial biopsies in order to direct therapeutic approaches. R2 measure could be used as a means to assess how directly
a drug targets a tumour type, i.e. whether a drug affects a tumour directly or via the micro-environment.
Below is a pictorial representation of examining neutral evolution with bulk sequencing.
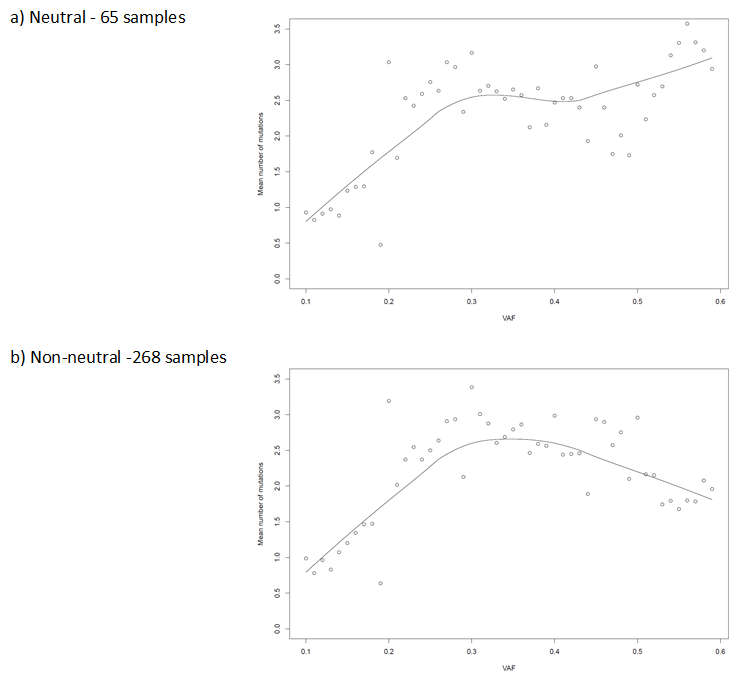
Distribution
In order to undertake this analysis, we used a standard sequencing pipeline for whole exomes. To assess R2 we calculated the gradient of the distribution of VAF for
cumulative mutations over time. Mutations, were considered, where there were more than 10 reads, with at least 3 mutations. And we only considered samples that had 12 private
mutations between a frequency of 0.12 and 2.4. We corrected for copy number at each mutation as adjusted and generated an R2 score.
It can be seen in the figure beneath when considering the distribution of sub-clonal mutations below 0.6, the relationship between the number of mutations and average variant
allele frequency (VAF) is linear in neutral samples indicating an absence of sub-clonity.
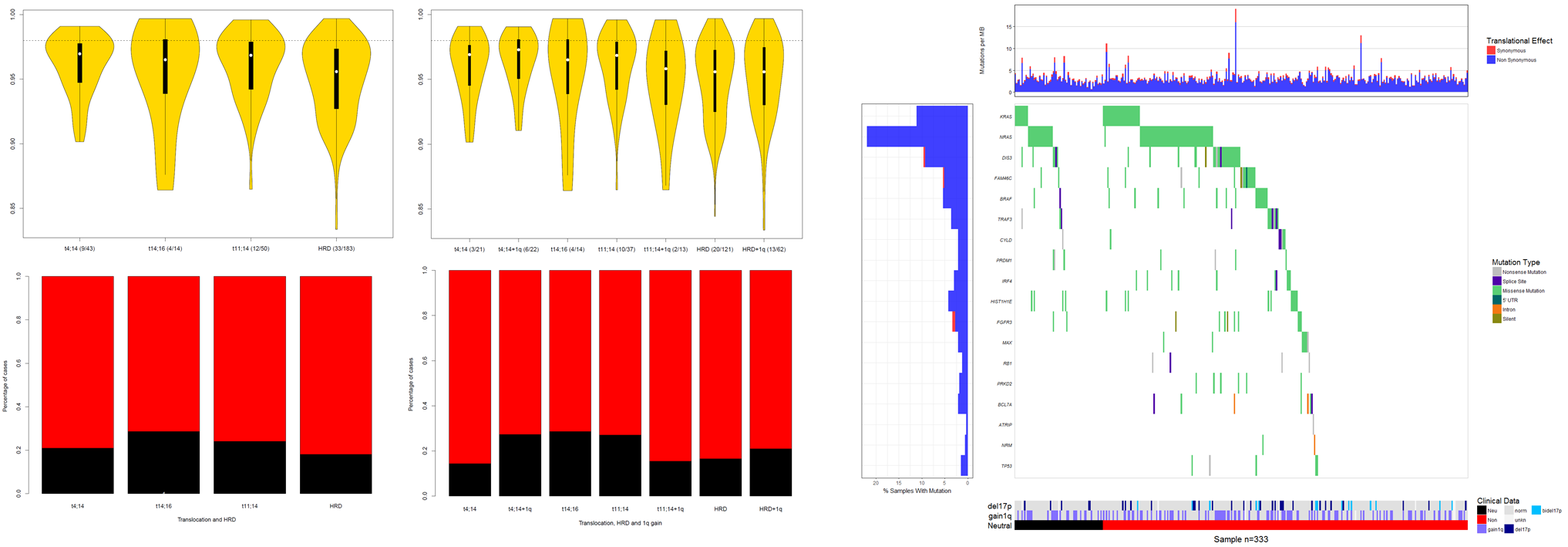
Subgroups and mutational signatures
As expected, a neutral tumour signature was seen to be enriched in myeloma subgroups (figure left), where disease transition has been shown to more rapid from the precursor MGUS disease.
The numbers were small but I was able to show that micro-environment enriched mutations were more prevalent in the non-neutral samples, indicating that these samples were
likely to be more micro-environment sensitive (figure right).
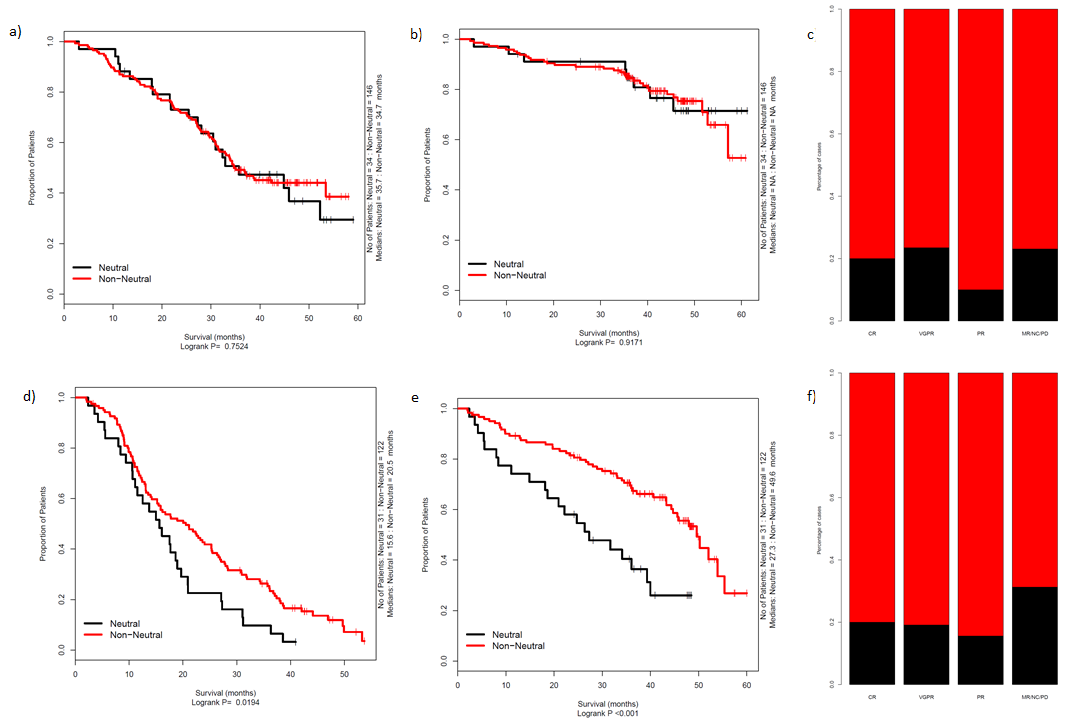
Survival and response
Indeed, when we compared OS and PFS in auto treated patients where the bulk of the tumour is removed we see little difference. Whilst in the non-intensively treated patient
received IMIDs, where the response to therapy is micro-environment dependent, we do see a difference in outcomes. Assessing independent data-sets from MMRF Commpass
cancer-register, we see a similar pattern.
Programs and packages
Survival analysis - survival, survivalminer, glmnet; Sequencing analysis - SnpEff, Onctotator, MutsigCV, GenVisR; Mutational signatures - deconstructSigs;
Data-visualisation - ggplot2, vioplot, matplotlib, seaborn.
References
◦ Neutral tumor evolution in myeloma is associated with poor prognosis. Johnson DC, Lenive O, Mitchell J, Jackson G, Owen R, Drayson M, Cook G, Jones JR, Pawlyn C, Davies FE, Walker BA, Wardell C, Gregory WM, Cairns D, Morgan GJ, Houlston RS, Kaiser MF. Blood. 2017 Oct 5;130(14):1639-1643. PMID: 28827410.
◦ Identification of neutral tumor evolution across cancer types. Williams MJ, Werner B, Barnes CP, Graham TA, Sottoriva A. Nature Genetics 2016, Mar;48(3):238-244. PMID: 26780609.
◦ Neutral tumor evolution? Tarabichi M, Martincorena I, Gerstung M, Leroi AM, Markowetz F et al. Nature Genetics 2018, Dec;50(12):1630-1633. PMID: 30374075.
◦ Reply to 'Neutral tumor evolution?' Heide T, Zapata L, Williams MJ, Werner B, Caravagna G, Barnes CP. Nature Genetics 2018, Dec;50(12):1633-1637. PMID: 30374073.