Myeloma GWAS
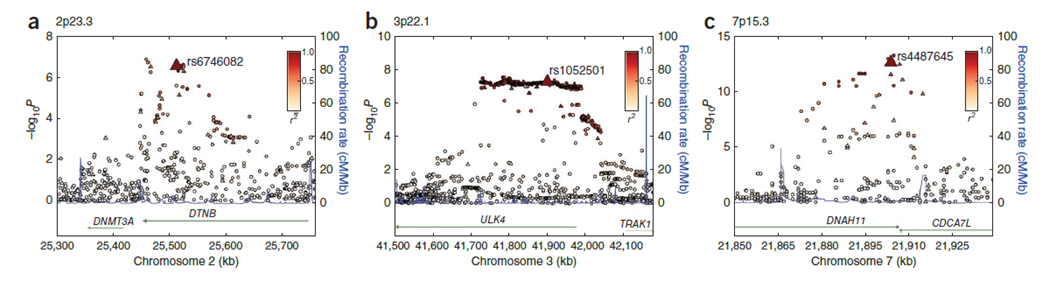
3p22.1 (ULK4) and 7p15.3 (CDC7AL)
There was much skepticism that a GWAS in myeloma would find any regions of association with risk. Myeloma is a late onset disease and thought to be initiated solely by random primary somatic
events. Although Myeloma families have been described, much of the epidemiological studies had focussed on toxic exposure rather than the influence of inherited factors,
see our review. In this first myeloma GWAS we performed an association analysis on 1,675 individuals with multiple
myeloma and 5,903 control subjects. I had previously used the myeloma case DNA on a custom SNP array, so I was able to select cases with good QC characteristics and
also able to filter for cases with non-European markers. Two genome-wide associations were found together with another third strong association proximal to DNMT3A.
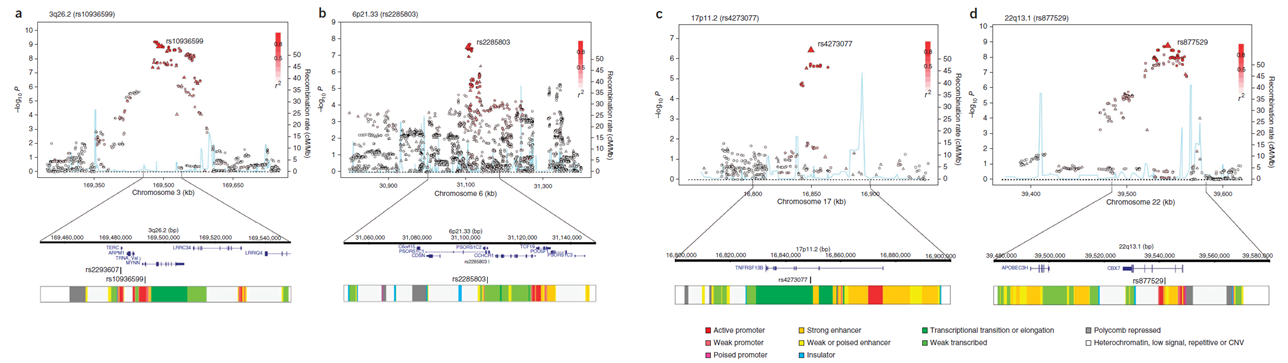
3q26.2 (TERC), 6p21.33 (PSORS1C2), 17p11.2 (TNFRSF13B) and 22q13.1 (CBX7)
In this second myeloma GWAS we were able to extend the dataset to 4,692 individuals with multiple myeloma (cases) and 10,990 controls. In this manuscript, we included the relationship
between SNP genotype and mRNA expression in CD138 selected plasma cells and lymphoblastoid cell lines. The genome-wide association plots are shown below.
6p22.3 (JARID2), 6q21 (ATG5), 7q36.1 (SMARCD3), 8q24.21 (CCAT1), 9p21.3 (CDKN2A), 10p12.1 (WAC), 16q23.1 (RFWD3) and 20q13.13 (PREX1).
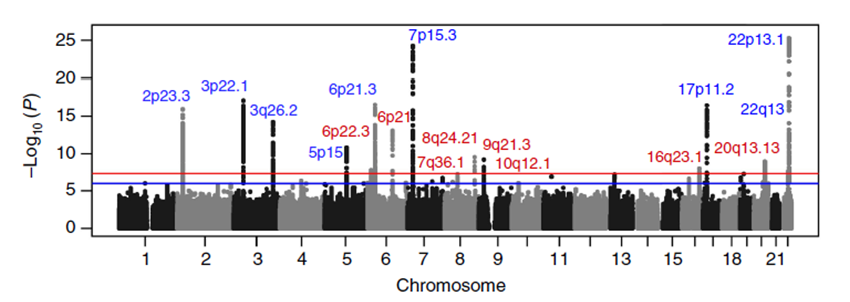
This manuscript contained an analysis of 9,866 cases and 239,188 controls, where we were able to take advantage of improved imputation methods and larger controls sets. A full eQTL analysis
was also included.The figure below is a Manhattan plot of myeloma risk associations.
Mendelian randomization
Mendelian randomization makes use of robustly linked allelic variants as instrumental variables(IVs). These robust IVs can be derived from independent studies.
IVs are then used to infer whether associations and a phenotype are causal. Mendelian randomization can avoid the confounding influence of environmental factors,
reflect life-long exposure and circumvent reverse causality.
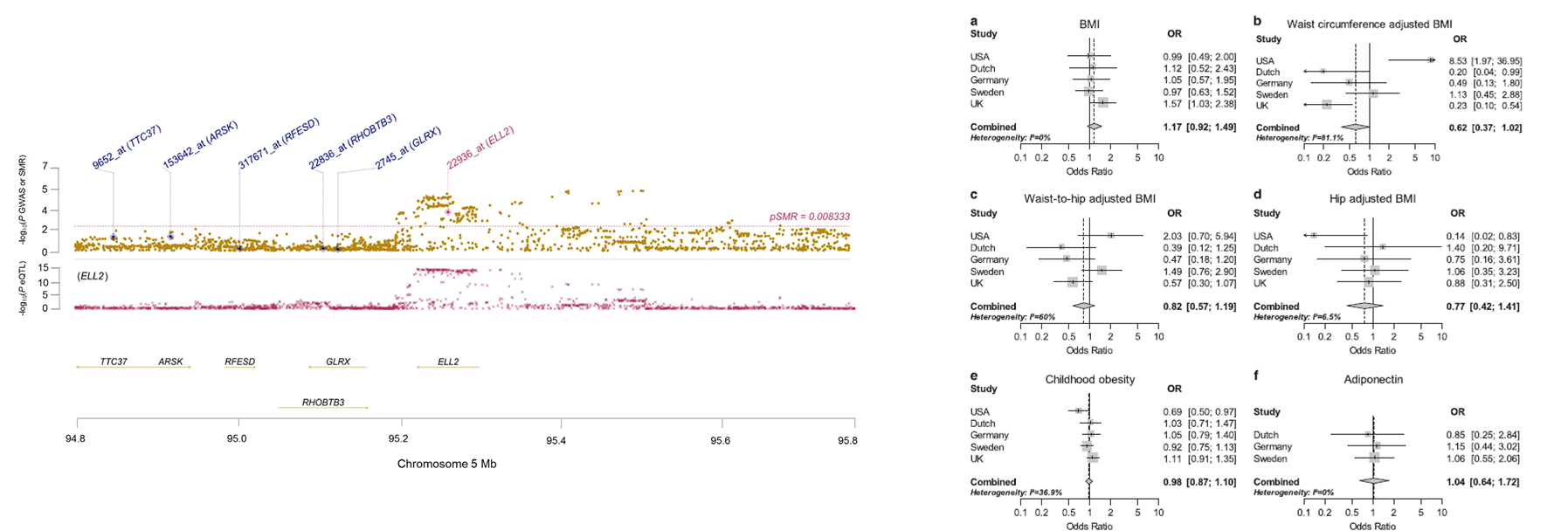
We have used mendelian randomization in combination with eQTL analysis provided evidence for differential ELL2 expression as being the basis of the
5q15 Myeloma risk association. I had previously performed an eQTL analysis in
841 Myeloma patients, and we used a summary data-based Mendelian randomization (SMR) analysis to test for pleiotropy between GWAS signal and cis-eQTL for genes within
1 Mb of the lead SNP to identify a causal relationship. We also used mendelian randomization to assess a previously described epidemiological link of obesity with
myeloma. Using genome-wide myeloma risk alleles as a proxy for life-long myeloma risk, together IVs for BMI, childhood obesity, adiponectin levels, we found no
support for link between myeloma risk and obesity.
On left is summary data-based mendelian randomization eQTL analysis at 5q15 and on the right are the meta-analysis odds ratios (OR) for myeloma per unit
increase in genetic risk score (s.d. trait) for each adiposity trait.
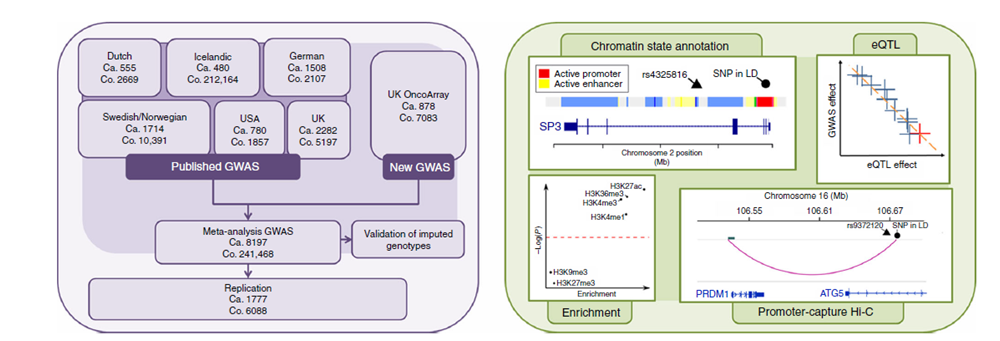
2q31.1 (SP3), 5q23.2 (CEP120), 7q22.3 (CCDC71L), 7q31.33 (POT1), 16p11.2 (SRCAP) and 19p13.11 (KLF2)
This study comprised of 9974 cases and 247,556 controls. This analysis included additional case and controls with the integration of information from gene expression (eQTL),
epigenetic profiling and in-situ Hi-C data. Below is an outline of the study numbers and experimental design
Pan Cancer GWAS
Following on from the initial myeloma GWAS, we were able to show the expected shared aetiology with a precursor disease stage
Monoclonal gammopathy of unknown significance. We went on to show a genetic correlation
between multiple myeloma and other B-cell disorders existed. Showing evidence for shared aetiology and pleiotropic risk loci with Hodgkin lymphoma and
chronic lymphocytic leukaemia
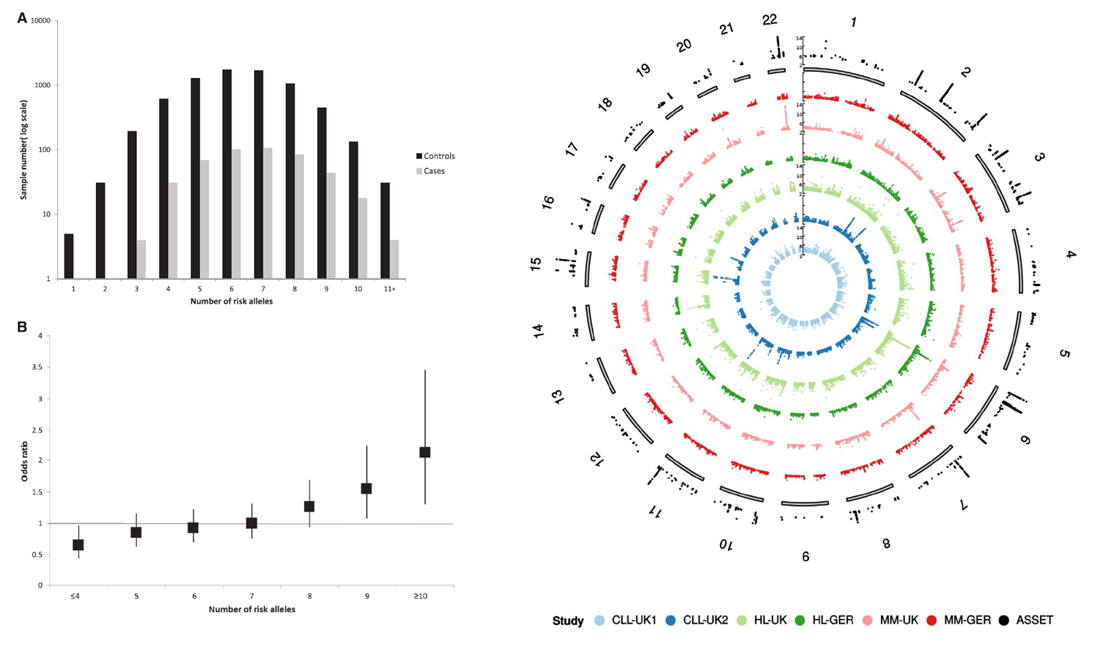
On the left is shown the proportion of MGUS cases and controls grouped according to the number of Myeloma risk alleles carried, and the increasing ORs for MGUS with increasing numbers of
myeloma risk alleles. On the right are Manhattan plots for each dataset by Chromosome.
Programs and packages
Association testing - PLINK versions 1.7-1.9, IMPUTE, EIGENSTRAT and SNPTEST; Meta-analysis - PLINK, META, METAL; Mendelinan randomisation - custom R script, MendelianRandomization v0.3.0, MeRP;
Data-visualisation - ggplot2, SNAP, circlize, matplotlib, seaborn,; HLA region analysis - SNP2HLA; Hi-C- HICUP and CHiCAGO; eQTL and meQTL - matrixQTL, FastQTL and PEER.
References
◦ Common variation at 3p22.1 and 7p15.3 influences multiple myeloma risk. Broderick P*, Chubb D*, Johnson DC*, Weinhold N, Försti A, Lloyd A, Olver B, Ma Y, Dobbins SE, Walker BA, Davies FE, Gregory WA, Childs JA, Ross FM, Jackson GH, Neben K, Jauch A, Hoffmann P, Mühleisen TW, Nöthen MM, Moebus S, Tomlinson IP, Goldschmidt H, Hemminki K, Morgan GJ, Houlston RS. Nat Genet. 2011 Nov 27;44(1):58-61. *These authors contributed equally to this work. PMID: 22120009
◦ Common variation at 3q26.2, 6p21.33, 17p11.2 and 22q13.1 influences multiple myeloma risk. Chubb D, Weinhold N, Broderick P, Chen B, Johnson DC, Försti A, Vijayakrishnan J, Migliorini G, Dobbins SE, Holroyd A, Hose D, Walker BA, Davies FE, Gregory WA, Jackson GH, Irving JA, Pratt G, Fegan C, Fenton JA, Neben K, Hoffmann P, Nöthen MM, Mühleisen TW, Eisele L, Ross FM, Straka C, Einsele H, Langer C, Dörner E, Allan JM, Jauch A, Morgan GJ, Hemminki K, Houlston RS, Goldschmidt H. Nat Genet. 2013 Oct;45(10):1221-1225. PMID: 23955597
◦ Genome-wide association study identifies multiple susceptibility loci for multiple myeloma. Mitchell JS, Li N, Weinhold N, Försti A, Ali M, van Duin M, Thorleifsson G, Johnson DC, Chen B, Halvarsson BM, Gudbjartsson DF, Kuiper R, Stephens OW, Bertsch U, Broderick P, Campo C, Einsele H, Gregory WA, Gullberg U, Henrion M, Hillengass J, Hoffmann P, Jackson GH, Johnsson E, Jöud M, Kristinsson SY, Lenhoff S, Lenive O, Mellqvist UH, Migliorini G, Nahi H, Nelander S, Nickel J, Nöthen MM, Rafnar T, Ross FM, da Silva Filho MI, Swaminathan B, Thomsen H, Turesson I, Vangsted A, Vogel U, Waage A, Walker BA, Wihlborg AK, Broyl A, Davies FE, Thorsteinsdottir U, Langer C, Hansson M, Kaiser M, Sonneveld P, Stefansson K, Morgan GJ, Goldschmidt H, Hemminki K, Nilsson B, Houlston RS. Nat Commun. 2016 Jul 1;7:12050. PMID: 27363682
◦ Genetic Predisposition to Multiple Myeloma at 5q15 Is Mediated by an ELL2 Enhancer Polymorphism. Li N, Johnson DC, Weinhold N, Kimber S, Dobbins SE, Mitchell JS, Kinnersley B, Sud A, Law PJ, Orlando G, Scales M, Wardell CP, Försti A, Hoang PH, Went M, Holroyd A, Hariri F, Pastinen T, Meissner T, Goldschmidt H, Hemminki K, Morgan GJ, Kaiser M, Houlston RS. Cell Rep. 2017 Sep 12;20(11):2556-2564. PMID: 28903037
◦ Assessing the effect of obesity-related traits on multiple myeloma using a Mendelian randomisation approach. Went M, Sud A, Law PJ, Johnson DC, Weinhold N, Försti A, van Duin M, Mitchell JS, Chen B, Kuiper R, Stephens OW, Bertsch U, Campo C, Einsele H, Gregory WM, Henrion M, Hillengass J, Hoffmann P, Jackson GH, Lenive O, Nickel J, Nöthen MM, da Silva Filho MI, Thomsen H, Walker BA, Broyl A, Davies FE, Langer C, Hansson M, Kaiser M, Sonneveld P, Goldschmidt H, Hemminki K, Nilsson B, Morgan GJ, Houlston RS. Blood Cancer J. 2017 Jun 16;7(6):e573. PMID: 28622301
◦ Identification of multiple risk loci and regulatory mechanisms influencing susceptibility to multiple myeloma. Went M, Sud A, Försti A, Halvarsson BM, Weinhold N, Kimber S, van Duin M, Thorleifsson G, Holroyd A, Johnson DC, Li N, Orlando G, Law PJ, Ali M, Chen B, Mitchell JS, Gudbjartsson DF, Kuiper R, Stephens OW, Bertsch U, Broderick P, Campo C, Bandapalli OR, Einsele H, Gregory WA, Gullberg U, Hillengass J, Hoffmann P, Jackson GH, Jöckel KH, Johnsson E, Kristinsson SY, Mellqvist UH, Nahi H, Easton D, Pharoah P, Dunning A, Peto J, Canzian F, Swerdlow A, Eeles RA, Kote-Jarai Z, Muir K, Pashayan N, Nickel J, Nöthen MM, Rafnar T, Ross FM, da Silva Filho MI, Thomsen H, Turesson I, Vangsted A, Andersen NF, Waage A, Walker BA, Wihlborg AK, Broyl A, Davies FE, Thorsteinsdottir U, Langer C, Hansson M, Goldschmidt H, Kaiser M, Sonneveld P, Stefansson K, Morgan GJ, Hemminki K, Nilsson B, Houlston RS; PRACTICAL consortium. Nat Commun. 2018 Sep 13;9(1):3707. PMID: 30213928
◦ Inherited genetic susceptibility to monoclonal gammopathy of unknown significance. Weinhold N, Johnson DC, Rawstron AC, Försti A, Doughty C, Vijayakrishnan J, Broderick P, Dahir NB, Begum DB, Hosking FJ, Yong K, Walker BA, Hoffmann P, Mühleisen TW, Langer C, Dörner E, Jöckel KH, Eisele L, Nöthen MM, Hose D, Davies FE, Goldschmidt H, Morgan GJ, Hemminki K, Houlston RS. Blood. 2014 Apr 17;123(16):2513-7. PMID: 24449210
◦ Genome-wide association analysis of chronic lymphocytic leukaemia, Hodgkin lymphoma and multiple myeloma identifies pleiotropic risk loci. Law PJ, Sud A, Mitchell JS, Henrion M, Orlando G, Lenive O, Broderick P, Speedy HE, Johnson DC, Kaiser M, Weinhold N, Cooke R, Sunter NJ, Jackson GH, Summerfield G, Harris RJ, Pettitt AR, Allsup DJ, Carmichael J, Bailey JR, Pratt G, Rahman T, Pepper C, Fegan C, von Strandmann EP, Engert A, Försti A, Chen B, Filho MI, Thomsen H, Hoffmann P, Noethen MM, Eisele L, Jöckel KH, Allan JM, Swerdlow AJ, Goldschmidt H, Catovsky D, Morgan GJ, Hemminki K, Houlston RS. Sci Rep. 2017 Jan 23;7:41071. PMID: 28112199
◦ Genetic correlation between multiple myeloma and chronic lymphocytic leukaemia provides evidence for shared aetiology. Went M, Sud A, Speedy H, Sunter NJ, Försti A, Law PJ, Johnson DC, Mirabella F, Holroyd A, Li N, Orlando G, Weinhold N, van Duin M, Chen B, Mitchell JS, Mansouri L, Juliusson G, Smedby KE, Jayne S, Majid A, Dearden C, Allsup DJ, Bailey JR, Pratt G, Pepper C, Fegan C, Rosenquist R, Kuiper R, Stephens OW, Bertsch U, Broderick P, Einsele H, Gregory WM, Hillengass J, Hoffmann P, Jackson GH, Jöckel KH, Nickel J, Nöthen MM, da Silva Filho MI, Thomsen H, Walker BA, Broyl A, Davies FE, Hansson M, Goldschmidt H, Dyer MJS, Kaiser M, Sonneveld P, Morgan GJ, Hemminki K, Nilsson B, Catovsky D, Allan JM, Houlston RS. Blood Cancer J. 2018 Dec 21;9(1):1. doi: 10.1038/s41408-018-0162-8. PMID: 30602759 PMID: 30373884