Admixture Genetics
Background
Admixture genetics has been of great personal interest to me as my family and I are very heterogeneous in terms of geographical ancestry and I wonder how the great breakthroughs
in genetics will translate to families such as mine.
Admixture studies are important because:
◦ Healthcare is important for all ancestral populations and therefore is essential we understand impact of treatments in individuals spanning all population groups. In the gnomAD exome and
genome database includes over 60% European sequences and less than 10% sequences from individuals of African ancestry at present.
◦ To accurately model genes and the influence on transcription and translation, it is important that we include as much information and variation as possible. Rare variant in one
population may be more common other populations, integrating eQTL and meQTL loci across populations for instance, can help build up phenotypic models.
◦ Ancestral populations differ due to differing selection pressure on populations in response to environmental pressure, these variates that differ between populations are also likely to be
enriched in causal variation.
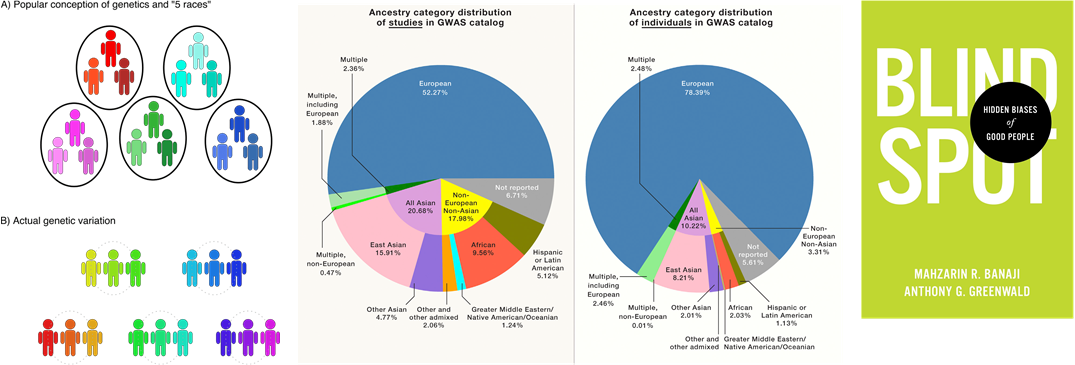
Left is an image from a Harvard review indicating the nature of population genetics, the center is a diagram from a cell review highlighting the disparity
in ancestry distribution in GWAS studies, right is a cover from the book of Blindspot: Hidden Biases of Good People. Given the disparity in the research community, we must continually
question the questions we are asking.
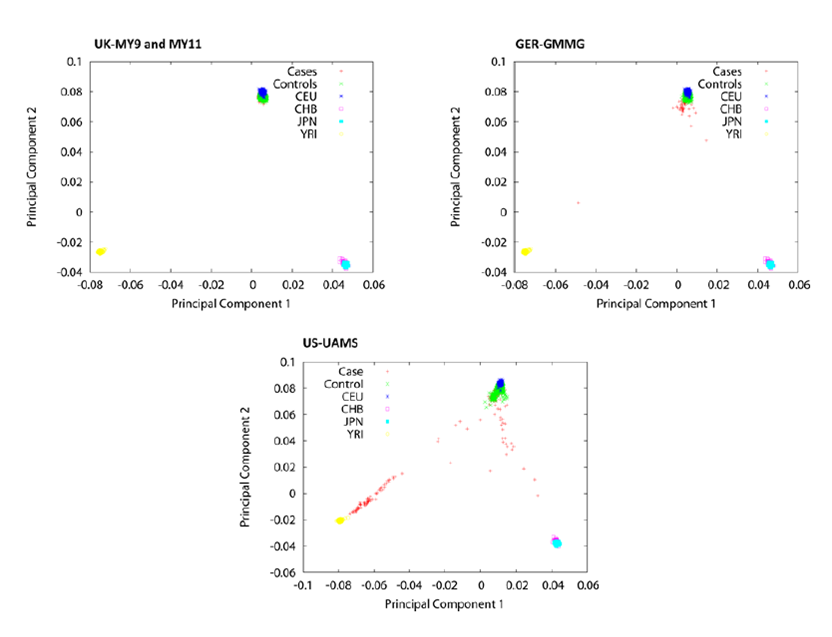
GWAS population outliers
All of my GWAS studies have been confined to european populations. In order to avoid inflating association test results due to an enrichment of non-european individuals in either of the comparator
groups, it has been necessary for me to identify non-europeans cases and excluded them from the analysis. Principle component analysis (PCA) analysis together with the
EIGENSTRAT correction method, allows us to identify population outliers and control for any inflation of test statistics by population
stratification using calculated eigenvectors.
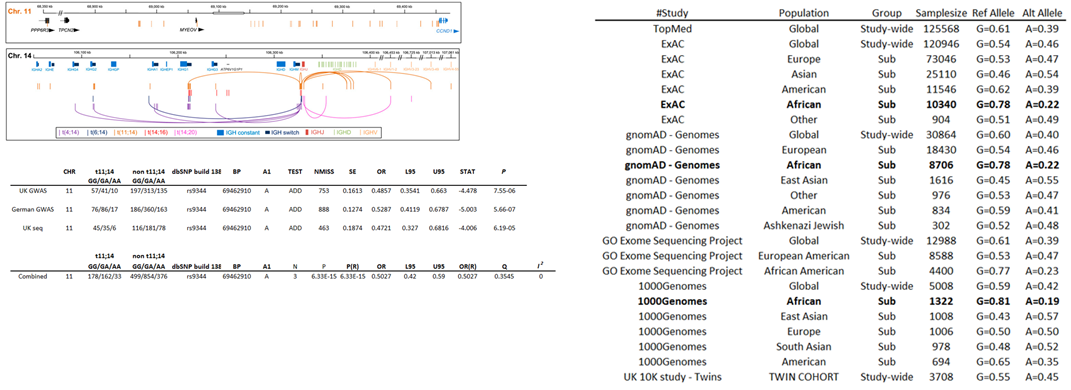
Ancestry variation and causal variates
A variant linked with the somatic cancer translocation t11;14 risk in myeloma is more prevalent in African populations. This variant is a well-described splice variant in
with results in short isoform of CCND1 which has an absent nucleus transporting motif, which leads to constitutive activation and increased proliferation. On the completion of our
study we internally speculated that population disparity of the variant may lead to an increase of t11;14 translocation in myeloma patients of African descent. Some support for
this hypothesis was provided in a 2018 study.
Programs and packages
Association testing - PLINK versions 1.7-1.9, IMPUTE, SNPTEST, EIGENSTRAT, Hail; Meta-analysis - PLINK, META, METAL; Mendelinan randomisation - custom R script, MendelianRandomization v0.3.0, MeRP;
Data-visualisation - ggplot2, SNAP, circlize, matplotlib, seaborn,; HLA region analysis - SNP2HLA; Hi-C- HICUP and CHiCAGO; eQTL and meQTL - matrixQTL, FastQTL and PEER.
References
◦ Genome-wide association study identifies variation at 6q25.1 associated with survival in multiple myeloma. Johnson DC, Weinhold N, Mitchell JS, Chen B, Kaiser M, Begum DB, Hillengass J, Bertsch U, Gregory WA, Cairns D, Jackson GH, Försti A, Nickel J, Hoffmann P, Nöethen MM, Stephens OW, Barlogie B, Davis FE, Hemminki K, Goldschmidt H, Houlston RS, Morgan GJ. Nat Commun. 2016 Jan 8;7:10290. PMID: 26743840
◦ Greenwald, A. G. & Banaji, M. R. (2013). Blindspot: Hidden Biases of Good People. Delacorte Press, ISBN 978-0553804645
◦ Trans-ethnic association study of blood pressure determinants in over 750,000 individuals. Giri A, Hellwege JN, Keaton JM, Park J, Qiu C, Warren HR et al. Nat Genet. 2019 PMID: 30578418 .
◦ The Missing Diversity in Human Genetic Studies.Sirugo, G., Williams, S. M., & Tishkoff, S. A. (2019). Cell, 177(1), 26–31. doi:10.1016/j.cell.2019.02.048 PMID: 30901543.
◦ Reply to 'Neutral tumor evolution?' Heide T, Zapata L, Williams MJ, Werner B, Caravagna G, Barnes CP. Nature Genetics 2018, Dec;50(12):1633-1637. PMID: 30374073.